Shortest-Path¶
Fix path and import caldera¶
from os.path import join, isfile
import os
import sys
def find_pkg(name: str, depth: int):
if depth <= 0:
ret = None
else:
d = ['..'] * depth
path_parts = d + [name, '__init__.py']
if isfile(join(*path_parts)):
ret = d
else:
ret = find_pkg(name, depth-1)
return ret
def find_and_ins_syspath(name: str, depth: int):
path_parts = find_pkg(name, depth)
if path_parts is None:
raise RuntimeError("Could not find {}. Try increasing depth.".format(name))
path = join(*path_parts)
if path not in sys.path:
sys.path.insert(0, path)
try:
import caldera
except ImportError:
find_and_ins_syspath('caldera', 3)
import caldera
Data Generation¶
To generate examples, we are going to use caldera’s networkx graph
generator utilties and the caldera.transforms.networkx module to
perform graph preprocessing.
import networkx as nx
import numpy as np
import pprint
import matplotlib.pyplot as plt
from caldera.testing import annotate_shortest_path
from caldera.transforms import Compose
from caldera.transforms.networkx import NetworkxAttachNumpyOneHot
from caldera.transforms.networkx import NetworkxNodesToStr
from caldera.transforms.networkx import NetworkxSetDefaultFeature
from caldera.transforms.networkx import NetworkxToDirected
from caldera.utils._tools import _resolve_range
from caldera.utils.nx.generators import _uuid_chain
from caldera.utils.nx.generators import chain_graph
from caldera.utils.nx.generators import compose_and_connect
from caldera.utils.nx.generators import random_graph
%matplotlib inline
def generate_shorest_path_example(n_nodes, density, path_length, compose_density = None):
d = _resolve_range(density)
l = _resolve_range(path_length)
if compose_density is None:
cd = d
else:
cd = _resolve_range(compose_density)
path = list(_uuid_chain(l))
h = chain_graph(path, nx.Graph)
g = random_graph(n_nodes, density=d)
graph = compose_and_connect(g, h, cd)
annotate_shortest_path(
graph,
True,
True,
source_key="source",
target_key="target",
path_key="shortest_path",
source=path[0],
target=path[-1],
)
preprocess = Compose(
[
NetworkxSetDefaultFeature(
node_default={"source": False, "target": False, "shortest_path": False},
edge_default={"shortest_path": False},
),
NetworkxAttachNumpyOneHot(
"node", "source", "_features", classes=[False, True]
),
NetworkxAttachNumpyOneHot(
"node", "target", "_features", classes=[False, True]
),
NetworkxAttachNumpyOneHot(
"edge", "shortest_path", "_target", classes=[False, True]
),
NetworkxAttachNumpyOneHot(
"node", "shortest_path", "_target", classes=[False, True]
),
NetworkxSetDefaultFeature(
node_default={"_features": np.array([0.0]), "_target": np.array([0.0])},
edge_default={"_features": np.array([0.0]), "_target": np.array([0.0])},
global_default={
"_features": np.array([0.0]),
"_target": np.array([0.0]),
},
),
NetworkxNodesToStr(),
NetworkxToDirected(),
]
)
return preprocess([graph])[0]
def draw_shortest_path(g, ax=None, node_size=10, edge_width=0.5, cmap=None, pos=None):
if ax is None:
fig = plt.figure(figsize=(3, 3))
ax = fig.gca()
ax.axis('off')
g = nx.to_undirected(g)
nodelist = list(g.nodes)
node_color = []
for n in nodelist:
node_color.append(g.nodes[n]['shortest_path'])
edge_list = []
edge_color = []
for n1, n2, edata in g.edges(data=True):
edge_list.append((n1, n2))
edge_color.append(edata['shortest_path'])
if cmap is None:
cmap = plt.get_cmap('seismic')
if pos is None:
pos = nx.layout.spring_layout(g)
elif callable(pos):
pos = pos(g)
nx.draw_networkx_nodes(g, pos=pos, node_color=node_color, node_size=node_size, ax=ax, cmap=cmap)
nx.draw_networkx_edges(g, pos=pos, width=edge_width, edge_color=edge_color, ax=ax, edge_cmap=cmap)
return ax
def custom_layout(g):
s1 = []
s2 = []
for n, ndata in g.nodes(data='shortest_path'):
if ndata:
s1.append(n)
else:
s2.append(n)
sg1 = g.subgraph(s1)
sg2 = g.subgraph(s2)
p1 = nx.layout.spring_layout(sg1)
p2 = nx.layout.spring_layout(sg2)
pos = {}
for p_ in [p1, p2]:
for n in p_:
pos[n] = p_[n]
return pos
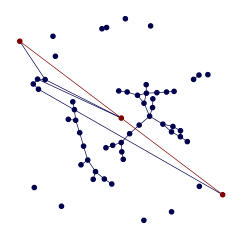
g = generate_shorest_path_example((10, 100), (0.03, 0.03), (5, 10), (0.01, 0.02))
draw_shortest_path(g, pos=custom_layout)
/home/justin/.cache/pypoetry/virtualenvs/caldera-fF8F2ZWq-py3.7/lib/python3.7/site-packages/traitlets/traitlets.py:2945: FutureWarning: --rc={'figure.dpi': 96} for dict-traits is deprecated in traitlets 5.0. You can pass --rc <key=value> ... multiple times to add items to a dict.
FutureWarning,
fig, axes = plt.subplots(3, 6, figsize=(12, 6))
for row in axes:
for ax in row:
g = generate_shorest_path_example((100, 150), (0.01, 0.03), (5, 30), (0.005, 0.02))
draw_shortest_path(g, ax, node_size=1, edge_width=0.2, pos=custom_layout)

We can convert networkx.Graph into GraphData
from caldera.data import GraphData
input_graph = GraphData.from_networkx(g, feature_key='_features')
target_graph = GraphData.from_networkx(g, feature_key='_target')
print(input_graph)
print(target_graph)
<GraphData size(n,e,g)=torch.Size([169, 450, 1]) features(n,e,g)=torch.Size([4, 1, 1])>
<GraphData size(n,e,g)=torch.Size([169, 450, 1]) features(n,e,g)=torch.Size([2, 2, 1])>
Creating Dataset¶
Generate networkx.Graphs, annotate the shortest path, convert
features to np.ndarray.
from caldera.dataset import GraphDataset
from caldera.data import GraphData, GraphBatch
from caldera.data import GraphDataLoader
from tqdm import tqdm
NUM_GRAPHS = 1000
NUM_NODES = (10, 100)
DENSITY = (0.01, 0.03)
PATH_LEN = (5, 10)
COMPOSITION_DENSITY = (0.01, 0.02)
nx_graphs = []
for _ in tqdm(range(NUM_GRAPHS)):
nxg = generate_shorest_path_example(NUM_NODES, DENSITY, PATH_LEN, COMPOSITION_DENSITY)
nx_graphs.append(nxg)
100%|██████████| 1000/1000 [00:03<00:00, 263.22it/s]
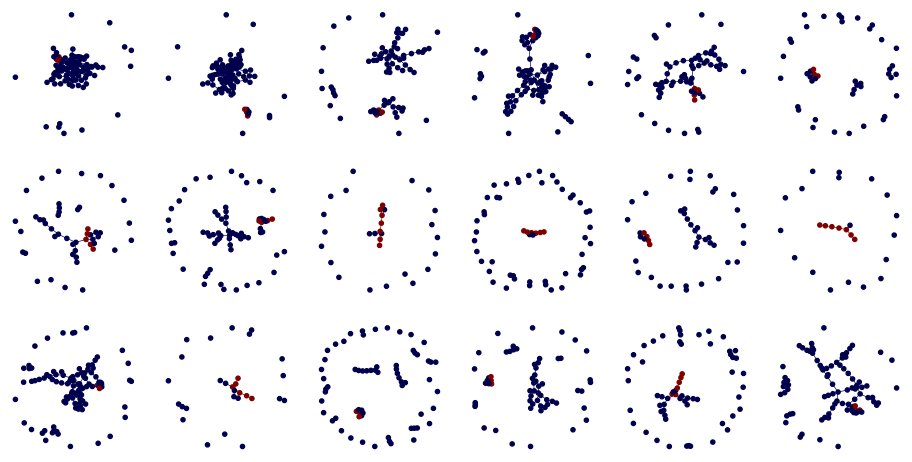
Check generated graphs.
from caldera.utils.functional import chain_each
fig, axes = plt.subplots(3, 6, figsize=(12, 6))
axes = chain_each()(axes)
for ax, g in zip(axes, nx_graphs):
draw_shortest_path(g, ax)
Create a paired data loader from input and target datalists.
input_datalist = [GraphData.from_networkx(g, feature_key='_features') for g in nx_graphs]
target_datalist = [GraphData.from_networkx(g, feature_key='_target') for g in nx_graphs]
loader = GraphDataLoader(input_datalist, target_datalist, batch_size=32)
for _input, _target in loader:
print(_input)
print(_target)
break
<GraphBatch size(n,e,g)=torch.Size([2118, 3444, 32]) features(n,e,g)=torch.Size([4, 1, 1])>
<GraphBatch size(n,e,g)=torch.Size([2118, 3444, 32]) features(n,e,g)=torch.Size([2, 2, 1])>
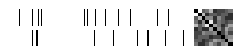
Plot tensor object fingerprints.
from caldera.data.utils import graph_matrix
from caldera.utils import functional
import seaborn as sns
def adj_fingerprint(data, step_size=50):
t = graph_matrix(data, include_edge_attr=False, fill_value=0, edge_value=1)
n = t.numpy().squeeze()
_n = np.zeros_like(n[:step_size, :step_size])
for i in np.arange(0, n.shape[0], step=step_size):
n2 = n[i:i+step_size, i:i+step_size]
pad = step_size - n2.shape[0]
if pad > 0:
n2 = np.hstack([n2, np.zeros((n2.shape[0], pad))])
n2 = np.vstack([n2, np.zeros((pad, n2.shape[1]))])
_n += n2
return _n
def plot_fingerprint(data):
fig = plt.figure(constrained_layout=True, figsize=(3, 0.5))
gs = fig.add_gridspec(2, 5)
ax1 = fig.add_subplot(gs[0, :-1])
ax2 = fig.add_subplot(gs[1, :-1])
ax3 = fig.add_subplot(gs[:, -1:])
sns.heatmap(data.x[:, :1].T, ax=ax1, cbar=False, xticklabels=False, yticklabels=False, cmap="gray")
sns.heatmap(data.e[:, :1].T, ax=ax2, cbar=False, xticklabels=False, yticklabels=False, cmap="gray")
sns.heatmap(adj_fingerprint(data, step_size=10), cbar=False, xticklabels=False, yticklabels=False, cmap="gray")
return fig
def split(n, step_size):
for i in np.arange(0, n.shape[0], step=step_size):
n2 = n[i:i+step_size, i:i+step_size]
pad = step_size - n2.shape[0]
if pad > 0:
n2 = np.hstack([n2, np.zeros((n2.shape[0], pad))])
n2 = np.vstack([n2, np.zeros((pad, n2.shape[1]))])
yield n2
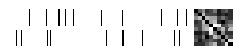
for _input, _target in functional.iter_count(3)(loader):
fig = plot_fingerprint(_target)
n = graph_matrix(_target).numpy().sum(axis=2)
x = np.hstack(list(split(n, 100)))
fig = plt.figure(figsize=(10, 2))
ax = fig.gca()
ax.set_title("Unrolled Adj Matrix")
sns.heatmap(x, ax=ax, cbar=False)
/home/justin/.cache/pypoetry/virtualenvs/caldera-fF8F2ZWq-py3.7/lib/python3.7/site-packages/seaborn/matrix.py:342: UserWarning: constrained_layout not applied. At least one axes collapsed to zero width or height.
ax.figure.draw(ax.figure.canvas.get_renderer())
/home/justin/.cache/pypoetry/virtualenvs/caldera-fF8F2ZWq-py3.7/lib/python3.7/site-packages/IPython/core/pylabtools.py:132: UserWarning: constrained_layout not applied. At least one axes collapsed to zero width or height.
fig.canvas.print_figure(bytes_io, **kw)

Notice that the input and target data are batched together according to
the batch size. Node attributes can be accessed via x, edge
attributes e and global attributes g. Now data is ready to be
loaded into a caldera network.
Graph Network¶
We are going to build a graph network to handle the data we just created.
We are going to use a flexible encoder -> core[x] -> decoder
architecture for this problem. The architecture consists of 4 main
networks, the encoder, core, decoder, and
output_transform networks. The encoder encodes graph data inputs
into arbitrary shapes. The core is the central graph message
processing network. The decoder decodes encoded data. Finally, the
output_transform transformed decoded data for final output.
Setting up the network looks like the following:
class Network(torch.nn.Module):
def __init__(...):
super().__init__()
self.config = {...}
self.encoder = ...
self.core = ...
self.decoder = ...
self.out_transform = ...
def forward(self, data, steps, save_all: bool = False):
"""The encoder -> core -> decode loop"""
encoded = self.encoder(data) # encode data
outputs = []
for _ in range(steps):
latent = self.core(encoded)
encoded = self.decoder(latent)
outputs.append(self.out_transform(latent)
return outputs
Flex Modules and Flex Dimensions¶
Setting up this network with the correct dimensions can become tricky,
so we introduce a new module, the Flex module, which can resolve
unknown dimensions on runtime. To make a module a Flex module, we
just call Flex with any torch.nn.Module, as in
Flex(torch.nn.Linear) or Flex(MyAwesomeModule). To initialize
the module with unknown dimensions, you use the flexible dimension
object Flex.d in places where the dimension is to be resolve on
runtime, as in Flex(torch.nn.Linear)(Flex.d(), 10).
from caldera.blocks import Flex
import torch
FlexLinear = Flex(torch.nn.Linear)
linear0 = torch.nn.Linear(3, 10)
flex_linear0 = FlexLinear(Flex.d(), 10)
print(linear0)
print(flex_linear0)
Linear(in_features=3, out_features=10, bias=True)
FlexBlock(
(unresolved_module): Linear(FlexDim(0, 1), 10,
)
Notice that the FlexBlock indicates it is current unresolved. To resovle it, we need to provide it with a data example. You’ll see the module is now resolved.
example = torch.zeros((1, 10))
flex_linear0(example)
print(flex_linear0)
FlexBlock(
(resolved_module): Linear(in_features=10, out_features=10, bias=True)
)
Aggregators¶
Aggregators are layers that indicate how data is processed and aggregated between neighbors.
Final Network¶
from caldera.blocks import NodeBlock, EdgeBlock, GlobalBlock
from caldera.blocks import AggregatingNodeBlock, AggregatingEdgeBlock, AggregatingGlobalBlock
from caldera.blocks import MultiAggregator
from caldera.blocks import Flex
from caldera.models import GraphCore, GraphEncoder
from caldera.defaults import CalderaDefaults as defaults
import torch
from caldera.defaults import CalderaDefaults as defaults
from caldera.blocks import Flex, NodeBlock, EdgeBlock, GlobalBlock, MLP, AggregatingEdgeBlock, AggregatingNodeBlock, \
MultiAggregator, AggregatingGlobalBlock
from caldera.models import GraphEncoder, GraphCore
from caldera.data import GraphBatch
class Network(torch.nn.Module):
def __init__(
self,
latent_sizes=(16, 16, 1),
out_sizes=(1, 1, 1),
latent_depths=(1, 1, 1),
dropout: float = None,
pass_global_to_edge: bool = True,
pass_global_to_node: bool = True,
activation=defaults.activation,
out_activation=defaults.activation,
edge_to_node_aggregators=tuple(["add", "max", "mean", "min"]),
edge_to_global_aggregators=tuple(["add", "max", "mean", "min"]),
node_to_global_aggregators=tuple(["add", "max", "mean", "min"]),
aggregator_activation=defaults.activation,
):
super().__init__()
self.config = {
"sizes": {
'latent': {
"edge": latent_sizes[0],
"node": latent_sizes[1],
"global": latent_sizes[2],
"edge_depth": latent_depths[0],
"node_depth": latent_depths[1],
"global_depth": latent_depths[2],
},
'out': {
'edge': out_sizes[0],
'node': out_sizes[1],
'global': out_sizes[2],
'activation': out_activation,
}
},
'activation': activation,
"dropout": dropout,
"node_block_aggregator": edge_to_node_aggregators,
"global_block_to_node_aggregator": node_to_global_aggregators,
"global_block_to_edge_aggregator": edge_to_global_aggregators,
"aggregator_activation": aggregator_activation,
"pass_global_to_edge": pass_global_to_edge,
"pass_global_to_node": pass_global_to_node,
}
###########################
# encoder
###########################
self.encoder = self._init_encoder()
self.core = self._init_core()
self.decoder = self._init_encoder()
self.output_transform = self._init_out_transform()
self.output_transform = GraphEncoder(
EdgeBlock(
torch.nn.Sequential(
Flex(torch.nn.Linear)(Flex.d(), 1), torch.nn.Sigmoid()
)
),
NodeBlock(
torch.nn.Sequential(
Flex(torch.nn.Linear)(Flex.d(), 1), torch.nn.Sigmoid()
)
),
GlobalBlock(Flex(torch.nn.Linear)(Flex.d(), 1)),
)
def _init_encoder(self):
return GraphEncoder(
EdgeBlock(Flex(MLP)(Flex.d(), self.config['sizes']['latent']['edge'], dropout=self.config['dropout'])),
NodeBlock(Flex(MLP)(Flex.d(), self.config['sizes']['latent']['node'], dropout=self.config['dropout'])),
GlobalBlock(Flex(MLP)(Flex.d(), self.config['sizes']['latent']['global'], dropout=self.config['dropout'])),
)
def _init_core(self):
edge_layers = [self.config['sizes']['latent']['edge']] * self.config['sizes']['latent']['edge_depth']
node_layers = [self.config['sizes']['latent']['node']] * self.config['sizes']['latent']['node_depth']
global_layers = [self.config['sizes']['latent']['global']] * self.config['sizes']['latent']['global_depth']
return GraphCore(
AggregatingEdgeBlock(
torch.nn.Sequential(
Flex(MLP)(Flex.d(), *edge_layers, dropout=self.config['dropout'], layer_norm=True),
)
),
AggregatingNodeBlock(
torch.nn.Sequential(
Flex(MLP)(Flex.d(), *node_layers, dropout=self.config['dropout'], layer_norm=True),
),
Flex(MultiAggregator)(
Flex.d(),
self.config["node_block_aggregator"],
activation=self.config["aggregator_activation"],
),
),
AggregatingGlobalBlock(
torch.nn.Sequential(
Flex(MLP)(
Flex.d(), *global_layers, dropout=self.config['dropout'], layer_norm=True
),
),
edge_aggregator=Flex(MultiAggregator)(
Flex.d(),
self.config["global_block_to_edge_aggregator"],
activation=self.config["aggregator_activation"],
),
node_aggregator=Flex(MultiAggregator)(
Flex.d(),
self.config["global_block_to_node_aggregator"],
activation=self.config["aggregator_activation"],
),
),
pass_global_to_edge=self.config["pass_global_to_edge"],
pass_global_to_node=self.config["pass_global_to_node"],
)
def _init_out_transform(self):
return GraphEncoder(
EdgeBlock(
torch.nn.Sequential(
Flex(torch.nn.Linear)(Flex.d(), self.config['sizes']['out']['edge']),
self.config['sizes']['out']['activation']()
)
),
NodeBlock(
torch.nn.Sequential(
Flex(torch.nn.Linear)(Flex.d(), self.config['sizes']['out']['node']),
self.config['sizes']['out']['activation']()
)
),
GlobalBlock(
torch.nn.Sequential(
Flex(torch.nn.Linear)(Flex.d(), self.config['sizes']['out']['global']),
self.config['sizes']['out']['activation']()
)
)
)
def _forward_encode(self, data):
e, x, g = self.encoder(data)
return GraphBatch(x, e, g, data.edges, data.node_idx, data.edge_idx)
def _forward_decode(self, data):
e, x, g = self.decoder(data)
return GraphBatch(x, e, g, data.edges, data.node_idx, data.edge_idx)
def _forward_core(self, latent0, data):
e = torch.cat([latent0.e, data.e], dim=1)
x = torch.cat([latent0.x, data.x], dim=1)
g = torch.cat([latent0.g, data.g], dim=1)
data = GraphBatch(x, e, g, data.edges, data.node_idx, data.edge_idx)
e, x, g = self.core(data)
return GraphBatch(x, e, g, data.edges, data.node_idx, data.edge_idx)
def _forward_out(self, data):
e, x, g = self.output_transform(data)
return GraphBatch(x, e, g, data.edges, data.node_idx, data.edge_idx)
def forward(self, data, steps, save_all: bool = False):
data = self._forward_encode(data)
latent0 = data
outputs = []
for _ in range(steps):
data = self._forward_core(latent0, data)
data = self._forward_decode(data)
out_data = self._forward_out(data)
if save_all:
outputs.append(out_data)
else:
outputs = [out_data]
return outputs
Provide example to resolve Flex modules
network = Network()
network.forward(_input, 10)
Training¶
# get device
if torch.cuda.is_available():
print("cuda available")
cuda_device = torch.cuda.current_device()
device = 'cuda:' + str(cuda_device)
else:
device = 'cpu'
# initialize network
network = Network()
# resolve
for input_batch, _ in loader:
x = input_batch.x
network(input_batch, 10)
break
# send to device
network.to(device, non_blocking=True)
loss_fn = torch.nn.BCELoss()
optimizer = torch.optim.AdamW(network.parameters())
# training loop
num_epochs = 10
running_loss = 0.
for epoch in tqdm(range(num_epochs)):
for input_batch, target_batch in loader:
network.train()
input_batch = input_batch.to(device)
target_batch = target_batch.to(device)
output = network(input_batch, 10)[0]
x, y = output.x, target_batch.x
loss = loss_fn(x.flatten(), y[:, 0].flatten())
loss.backward()
optimizer.step()
optimizer.zero_grad()
network.eval()
100%|██████████| 10/10 [00:31<00:00, 3.13s/it]